biotechrabbit™ Pfu PCR Master Mix is a perfect choice for fast, high-fidelity PCR setup that reduces the time required for calculation and pipetting and eliminates the need for buffer optimization. It is designed for routine high-throughput, high-fidelity amplification of targets up to 3–4 kb in size.
The 2X Pfu PCR Master Mix contains Pfu DNA Polymerase, extremely high-quality dNTPs and optimized PCR buffer; thus, only template, PCR primers and PCR-grade water are added. For the most demanding applications, the supplied 5× PCR Enhancer can be optionally be used to improve results when using templates with GC-rich sequences and complex structures.
Pfu DNA Polymerase exhibits approximately 10 times higher accuracy compared to Taq DNA polymerase. Pfu DNA Polymerase produces blunt-end PCR products suitable for blunt cloning.
|
Component |
Composition |
|
Pfu PCR Master Mix |
Optimized 2X Pfu PCR Master Mix |
|
5× PCR Enhancer |
Proprietary PCR enhancer mix |
|
STORAGE |
−20°C (until expiry date – see product label) |
Quality Control
Functional assay
Human genomic DNA was amplified using the Pfu PCR Master Mix and specific primers to produce a distinct band.
Prevention of PCR contamination
When assembling the amplification reactions, care should be taken to eliminate the possibility of contamination with undesired DNA.
- Use separate clean areas for preparation of samples and reaction mixtures and for cycling.
- Wear fresh gloves. Use sterile tubes and pipette tips with aerosol filters for PCR setup.
- Use only water and reagents that are free of DNA and nucleases.
- With every PCR setup, perform a contamination control reaction that does not include template DNA.
Standard PCR setup
The standard PCR protocol using biotechrabbit reaction buffer provides excellent results for most applications. Optimization might be necessary for certain conditions, such as the amplification of long targets, high GC or AT content, strong template secondary structures or insufficient template purity. In such cases, optimization of template purification (see biotechrabbit nucleic acid purification kits), primer design and annealing temperature is recommended.
The best conditions for each primer-template can be optimized with the following:
- Choosing the optimal quantities of template and primers
- Using a PCR Enhancer (i.e. BR1900201) for low amounts of template, impure or GC-rich templates
- Optimizing cycling conditions
Basic Protocol
- The Master Mix is designed to be used without any optimization as it has all necessary reaction components in optimal amounts for successful PCR.
- Optionally, use the supplied 5× PCR Enhancer to increase the yield and to lower the background in more complicated PCR reactions (low amounts of template, impure or GC-rich template).
- Thaw on ice and mix all reagents well.
- Keep all reagents and reactions on ice.
- Pipet the master mix into thin-walled 0.2 ml PCR tubes.
- Add template and primers separately if they are not used in all reactions.
|
Component |
Volume |
Final concentration |
|
Pfu PCR Master Mix, 2× |
25 µl |
1× |
|
5× PCR Enhancer (optional - BR1900201) |
10 µl |
1× |
|
Forward primer |
Variable |
0.2–1 µM |
|
Reverse primer |
Variable |
0.2–1 µM |
|
Template DNA |
Variable |
10 pg–1 μg |
|
|
Use 0.01–1 ng for plasmid or phage DNA and 0.1–1 μg for genomic DNA |
|
|
Nuclease free water |
Variable |
|
|
Total volume |
50 µl |
|
- Mix and centrifuge briefly to collect the liquid in the bottom of the tube.
- Place in the PCR cycler.
Cycling Program
|
Step |
Temperature |
Time |
Cycles |
|
Initial activation |
95°C |
2 min |
1 |
|
Denaturation |
95°C |
30 s |
25–35 |
|
Annealing |
55°C |
30–45 s |
25–35 |
|
|
Approximately 5°C below Tm of primers |
||
|
Extension |
72°C |
2 min/kb |
25–35 |
|
Final extension |
72°C |
5 min |
1 |
|
|
To extend all incomplete PCR products |
||
|
Storage in the cycler |
4°C |
Indefinitely |
1 |
- Add loading dye solution (see DNA Loading Dye, 6×, cat. no. BR0800301) to the reactions to analyze PCR products on a gel or store them at −20°C.
- For cloning, always purify the PCR product from a gel (see BR0700401 GenUP™ Gel Extraction Kit).
You may also be interested in the following product(s)
RTS 100 E. coli HY Kit
Package Sizes |
GenUP™ FFPE Paraffin Removal Solution
Package Sizes |
4X CAPITAL™ qPCR Green Master Mix
Package Sizes |
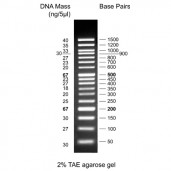
50 bp DNA Ladder, ready to use
Package Sizes |
UPstart™ Plus Taq Antibody, 1 mg/ml
Package Sizes |
